带有数据增强的 COCO 读取器#
这是一个示例,演示了如何将 COCO 读取器(从 COCO 数据集加载数据)与图像检测和分割用例中使用的一些典型数据增强方法结合使用。COCO 数据集包含一个图像目录和一个注释文件,其中包含有关边界框、标签和分割掩码的信息。
[1]:
from nvidia.dali.pipeline import Pipeline
import nvidia.dali.types as types
import nvidia.dali.fn as fn
import nvidia.dali.math as math
import numpy as np
from time import time
import os.path
import random
random.seed(1231231) # Random is used to pick colors
test_data_root = os.environ["DALI_EXTRA_PATH"]
file_root = os.path.join(test_data_root, "db", "coco", "images")
annotations_file = os.path.join(test_data_root, "db", "coco", "instances.json")
num_gpus = 1 # Single GPU for this example
device_id = 0
batch_size = 32
num_threads = 4 # Number of CPU threads
让我们从定义一个仅加载数据的简单 pipeline 开始
[2]:
pipe = Pipeline(
batch_size=batch_size, num_threads=num_threads, device_id=device_id
)
with pipe:
inputs, bboxes, labels, polygons, vertices = fn.readers.coco(
file_root=file_root,
annotations_file=annotations_file,
polygon_masks=True, # Load segmentation mask data as polygons
# Bounding boxes to be expressed as left, top, right, bottom coordinates
ltrb=True,
)
images = fn.decoders.image(inputs, device="mixed")
pipe.set_outputs(images, bboxes, labels, polygons, vertices)
pipe.build()
我们现在可以运行 pipeline 并可视化结果
[3]:
import matplotlib.pyplot as plt
import matplotlib.patches as patches
def plot_coco_sample(
image, bboxes, labels, mask_polygons, mask_vertices, relative_coords=False
):
H, W = image.shape[0], image.shape[1]
fig, ax = plt.subplots(dpi=160)
# Displaying the image
ax.imshow(image)
# Bounding boxes
for bbox, label in zip(bboxes, labels):
l, t, r, b = bbox * [W, H, W, H] if relative_coords else bbox
rect = patches.Rectangle(
(l, t),
width=(r - l),
height=(b - t),
linewidth=1,
edgecolor="#76b900",
facecolor="none",
)
ax.add_patch(rect)
# Segmentation masks
for polygon in mask_polygons:
mask_idx, start_vertex, end_vertex = polygon
# Select polygon vertices
polygon_vertices = mask_vertices[start_vertex:end_vertex]
# Scale relative coordinates to the image dimensions, if necessary
polygon_vertices = (
polygon_vertices * [W, H] if relative_coords else polygon_vertices
)
poly = patches.Polygon(
polygon_vertices, True, facecolor="#76b900", alpha=0.7
)
ax.add_patch(poly)
plt.show()
def show(outputs, relative_coords=False):
i = 16 # Picked a sample idx that shows more than one bounding box
images, bboxes, labels, mask_polygons, mask_vertices = outputs
plot_coco_sample(
images.as_cpu().at(i),
bboxes.at(i),
labels.at(i),
mask_polygons.at(i),
mask_vertices.at(i),
relative_coords=relative_coords,
)
outputs = pipe.run()
show(outputs)

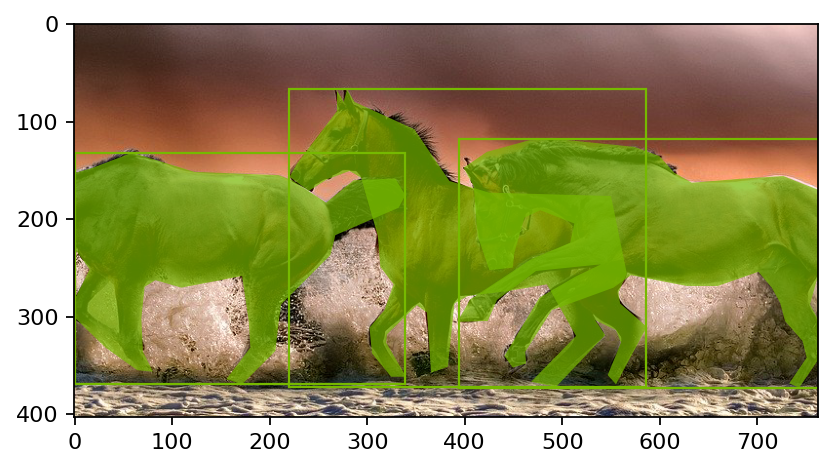
在检测和分割用例中应用的一种典型数据增强方法是对图像进行随机裁剪,并限制裁剪后的图像中至少存在一个 ground truth 框。在 DALI 中,我们为此使用 RandomBBoxCrop。RandomBBoxCrop 算子将边界框和与其关联的标签以及裁剪操作的一组约束作为输入。结果是裁剪窗口的锚点和形状,以及处理后的边界框和标签。以相对坐标表示的锚点和形状输出可以直接馈送到 DALI 的 Slice 算子,以提取图像的感兴趣区域。输出的边界框和标签经过处理,仅包含裁剪窗口内的那些,并且坐标映射到新的坐标空间。RandomBBoxCrop 不处理分割掩码,因此掩码坐标需要单独映射到新的坐标空间。
[4]:
# Wrapping the pipeline definition in separate functions that we can reuse later
def coco_reader_def():
inputs, bboxes, labels, polygons, vertices = fn.readers.coco(
file_root=file_root,
annotations_file=annotations_file,
polygon_masks=True, # Load segmentation mask data as polygons
# Bounding box and mask polygons to be expressed in relative coordinates
ratio=True,
# Bounding boxes to be expressed as left, top, right, bottom coordinates
ltrb=True,
)
return inputs, bboxes, labels, polygons, vertices
def random_bbox_crop_def(bboxes, labels, polygons, vertices):
# RandomBBoxCrop works with relative coordinates
# The arguments have been selected to produce a significantly visible crop
# To learn about all the available options, see the documentation
anchor_rel, shape_rel, bboxes, labels, bbox_indices = fn.random_bbox_crop(
bboxes,
labels,
aspect_ratio=[0.5, 2], # Range of aspect ratios
# No minimum intersection-over-union, for demo purposes
thresholds=[0.0],
allow_no_crop=False, # No-crop is disallowed, for demo purposes
# Scale range of the crop with respect to the image shape
scaling=[0.3, 0.6],
seed=12345, # Fixed random seed for deterministic results
bbox_layout="xyXY", # left, top, right, back
output_bbox_indices=True, # Output indices of the filtered bounding boxes
)
# Select mask polygons of those bounding boxes that remained in the image
polygons, vertices = fn.segmentation.select_masks(
bbox_indices, polygons, vertices
)
return anchor_rel, shape_rel, bboxes, labels, polygons, vertices
pipe = Pipeline(
batch_size=batch_size,
num_threads=num_threads,
exec_dynamic=True,
device_id=device_id,
)
with pipe:
inputs, bboxes, labels, polygons, vertices = coco_reader_def()
anchor_rel, shape_rel, bboxes, labels, polygons, vertices = (
random_bbox_crop_def(bboxes, labels, polygons, vertices)
)
# Partial decoding of the image
images = fn.decoders.image_slice(
inputs,
anchor_rel,
shape_rel,
normalized_anchor=True,
normalized_shape=True,
device="mixed",
)
# Cropped image dimensions
crop_shape = images.shape(dtype=types.FLOAT) # HWC
crop_h = crop_shape[0]
crop_w = crop_shape[1]
# Adjust masks coordinates to the coordinate space of the cropped image,
# while also converting relative to absolute coordinates by mapping
# the top-left corner (anchor_rel_x, anchor_rel_y), to (0, 0)
# and the bottom-right corner (anchor_rel_x+shape_rel_x,
# anchor_rel_y+shape_rel_y) to (crop_w, crop_h)
MT_vertices = fn.transforms.crop(
from_start=anchor_rel,
from_end=(anchor_rel + shape_rel),
to_start=(0.0, 0.0),
to_end=fn.stack(crop_w, crop_h),
)
vertices = fn.coord_transform(vertices, MT=MT_vertices)
# Convert bounding boxes to absolute coordinates
MT_bboxes = fn.transforms.crop(
to_start=(0.0, 0.0, 0.0, 0.0),
to_end=fn.stack(crop_w, crop_h, crop_w, crop_h),
)
bboxes = fn.coord_transform(bboxes, MT=MT_bboxes)
pipe.set_outputs(images, bboxes, labels, polygons, vertices)
pipe.build()
outputs = pipe.run()
show(outputs)
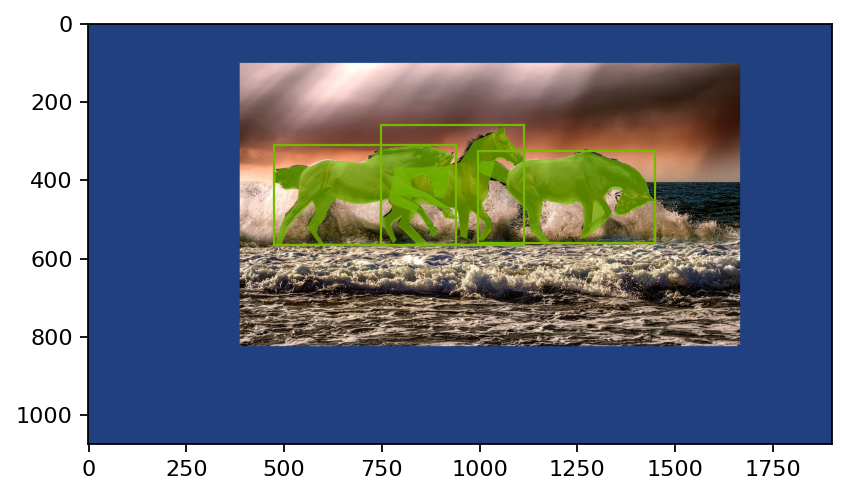
在下面的 pipeline 中,我们将图像粘贴到更大的画布上并水平翻转
[5]:
pipe = Pipeline(
batch_size=batch_size,
num_threads=num_threads,
device_id=device_id,
exec_dynamic=True,
seed=43210,
)
with pipe:
inputs, bboxes, labels, polygons, vertices = coco_reader_def()
images = fn.decoders.image(inputs)
orig_shape = images.shape()
images = images.gpu()
px = fn.random.uniform(range=(0, 1))
py = fn.random.uniform(range=(0, 1))
ratio = fn.random.uniform(range=(1, 2))
images = fn.paste(
images, paste_x=px, paste_y=py, ratio=ratio, fill_value=(32, 64, 128)
)
bboxes = fn.bbox_paste(
bboxes, paste_x=px, paste_y=py, ratio=ratio, ltrb=True
)
scale = 1.0 / ratio
margin = ratio - 1.0
pxy = fn.stack(px, py)
pxy = scale * pxy * margin
vertices = scale * vertices + pxy
# 100% probability for demo purposes
should_flip = fn.random.coin_flip(probability=1.0)
images = fn.flip(images, horizontal=should_flip)
bboxes = fn.bb_flip(bboxes, horizontal=should_flip, ltrb=True)
vertices = fn.coord_flip(vertices, flip_x=should_flip)
pipe.set_outputs(images, bboxes, labels, polygons, vertices)
pipe.build()
outputs = pipe.run()
show(outputs, relative_coords=True)
在某些分割和检测数据集中,对象的密度非常低。这可能会导致在随机裁剪图像时,背景在训练过程中被过度表示。为了弥补这一点,我们可以选择以一定概率将裁剪窗口居中于前景像素。
[6]:
pipe = Pipeline(
batch_size=batch_size,
num_threads=num_threads,
device_id=device_id,
seed=12345,
)
with pipe:
# COCO reader, with piwelwise masks
inputs, bboxes, labels, masks = fn.readers.coco(
file_root=file_root,
annotations_file=annotations_file,
pixelwise_masks=True, # Load segmentation pixelwise mask data
)
images = fn.decoders.image(inputs)
# COCO reader produces three dimensions (H, W, 1). Here we are just removing
# the trailing dimension rel_shape=(1, 1) means keep the first two dimensions
# as they are.
masks = fn.reshape(masks, rel_shape=(1, 1))
# Select random foreground pixels with 70% probability and random pixels
# with 30% probability
# Foreground pixels are by default those with value higher than 0.
center = fn.segmentation.random_mask_pixel(
masks, foreground=fn.random.coin_flip(probability=0.7)
)
# Random crop shape (can also be constant)
crop_w = fn.random.uniform(range=(200, 300), dtype=types.INT64)
crop_h = fn.random.uniform(range=(200, 300), dtype=types.INT64)
crop_shape = fn.stack(crop_h, crop_w)
# Calculating anchor for slice (top-left corner of the cropping window)
crop_anchor = center - crop_shape // 2
# Slicing image and mask.
# Note that we are allowing padding when sampling out of bounds, since
# a foreground pixel can appear near the edge of the image.
out_image = fn.slice(
images,
crop_anchor,
crop_shape,
axis_names="HW",
out_of_bounds_policy="pad",
)
out_mask = fn.slice(
masks,
crop_anchor,
crop_shape,
axis_names="HW",
out_of_bounds_policy="pad",
)
pipe.set_outputs(
images, masks, center, crop_anchor, crop_shape, out_image, out_mask
)
pipe.build()
outputs = pipe.run()
i = 16
image = outputs[0].at(i)
mask = outputs[1].at(i)
center = outputs[2].at(i)
anchor = outputs[3].at(i)
shape = outputs[4].at(i)
out_image = outputs[5].at(i)
out_mask = outputs[6].at(i)
fig, ax = plt.subplots(dpi=160)
ax.imshow(image)
ax.imshow(mask, cmap="jet", alpha=0.5)
rect = patches.Rectangle(
(anchor[1], anchor[0]),
width=shape[1],
height=shape[0],
linewidth=1,
edgecolor="#76b900",
facecolor="none",
)
ax.add_patch(rect)
ax.scatter(center[1], center[0], s=10, edgecolor="#76b900")
plt.title("Original Image/Mask with random crop window and center")
plt.show()
fig, ax = plt.subplots(dpi=160)
ax.imshow(out_image)
ax.imshow(out_mask, cmap="jet", alpha=0.5)
plt.title("Cropped Image/Mask")
plt.show()

